G protein coupled receptors (GPCRs) are the largest family of membrane receptor proteins in the human body, playing important roles in cellular signaling and closely related to human diseases. They are the target of over 40% of marketed drugs. According to the differences in structure and sequence, GPCRs in the human body can be divided into four types: A, B, C, and F. In recent years, structural and functional studies have revealed the activation mechanisms of GPCRs of classes A, B1, B2, and F that function in monomeric form. Research has shown that monomeric GPCRs coupled to G proteins follow a similar pattern, where receptor activation causes the outward migration of the transmembrane domain TM6, resulting in the formation of larger pockets for binding to the C-terminus of the G α subunit, thereby activating downstream signaling pathways of the G protein. However, for a long time, the molecular mechanism of C-class GPCR coupled with G protein, which functions in a dimeric form, has not been revealed, seriously hindering people's understanding of the signal transduction mechanism of C-class GPCR.
On April 28, 2021, Professor Jianfeng Liu's team from the School of Life Sciences and Technology and the Key Laboratory of Molecular Biophysics of the Ministry of Education at Huazhong University of Science and Technology, together with Professor Yan Zhang's team from the School of Medicine at Zhejiang University and the Liangzhu Laboratory, published a research paper titled "Structural basis of GABAB receptor Gi protein coupling" online in the journal Nature. This study breakthrough identified the high-resolution cryo electron microscopy structure of the heterodimeric GABAB receptor G protein complex, revealing for the first time the unique mode of asymmetric activation of G protein by dimeric GPCRs.
C-class GPCRs mainly include metabolic gamma aminobutyric acid receptors (GABAB), metabolic glutamate receptors (mGlu), calcium sensitive receptors (CaSR), and taste receptor 1 (Taste 1). Class C receptors are associated with numerous neurological and psychiatric disorders, including epilepsy, pain, anxiety, depression, schizophrenia, drug addiction, Rett syndrome, and epileptic encephalopathy. GABAB receptor is the first discovered and proven GPCR heterodimer, and is a typical representative of C-class GPCR dimers. GABAB receptors are widely expressed in the central nervous system and are involved in important physiological processes such as learning, memory, and synaptic signaling. GABAB receptors are composed of two subunits, GB1 and GB2, each consisting of an extracellular domain (VFT), a seven transmembrane domain (TMD), and an intracellular domain. GABAB receptor activation adopts a typical asymmetric regulatory mechanism, where the VFT of GB1 is responsible for ligand recognition, while the TMD coupled Gi/o protein of GB2 regulates downstream signals.
The collaborative team of Liu Jianfeng and Zhang Yan first identified the fine three-dimensional spatial structure of the inactive and active states of full-length GABAB receptors internationally in 2020, elucidated the conformational transition process during GABAB receptor activation, identified the binding pocket of positive allosteric modulators, and greatly promoted the research on the activation mechanism of GABAB receptors and the development of targeted drugs. At the same time, this work also analyzed for the first time the structure of low resolution GABAB-Gi complexes and found that due to potential steric hindrance, dimerized GABAB receptors can only couple to one G protein. However, the molecular mechanism of its asymmetric activation and signal transduction coupled with G proteins has not yet been revealed.
On the basis of previous work, the research team conducted further research on the topic, overcoming challenges such as complex assembly, uniform frozen sample preparation, and dynamic conformation data processing, and finally completed the electron microscopy structural identification of high-resolution GABAB-Gi protein complex (Figure 1a). Structural analysis revealed that, unlike monomeric GPCRs, when GABAB receptor couples with G protein, both TM6 subunits of the transmembrane domain did not undergo outward migration, but TM3 and TM5 subunits of GB2 showed smaller outward expansion, resulting in a shallow pocket binding G protein in the three intracellular loops. Compared with other structures that bind to monomeric GPCRs and G proteins, the insertion depth of the G protein α 5 helix into GABAB receptors is about 10 Å lower, and it mainly interacts with the intracellular loop. These results, combined with extensive functional testing validation and previous functional studies of other C-class GPCRs, suggest that the C-class GPCR dimers all adopt a unique G protein coupling mode that differs from the activation of A, B1, B2, and F-class GPCRs (Figure 1b-d).
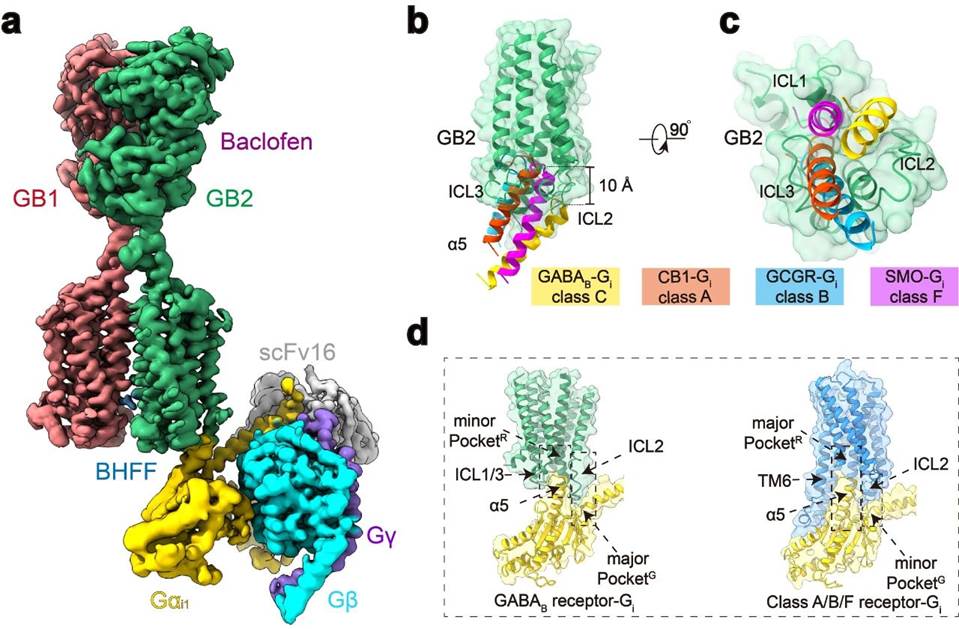
Figure 1: Novel structure of GABAB conjugated G protein complex
In addition, the study found fine conformational changes in GABAB coupled with G protein, including rotation between GB1 and GB2, as well as detailed changes in the TMD of GB2 subunit. Through structural analysis, researchers found that after binding GABAB receptors to G proteins with agonists, GB2-TMD further rotates counterclockwise compared to GB1-TMD, bringing the transmembrane domain interface of GB1 and GB2 closer and stabilizing the activated state. Researchers focused on the fine conformational changes within GB2-TMD and found that the conformational changes at the intracellular end of GB2 TM3/TM5 originate from the large angle rotation of the F586 side chain benzene ring on TM3. Functional experimental analysis found that the ability of receptor coupled G protein significantly decreased after F586 was mutated into small side chain alanine. In addition, at the bottom of GB2-TMD, salt bridges formed by multiple charged amino acids further stabilize the activated state of the receptor, revealing the structural basis of GB2 subunit binding to G protein (Figure 2). The unique conformational changes of GB2 subunits ultimately determine the asymmetric activation mode of GABAB coupled G proteins.
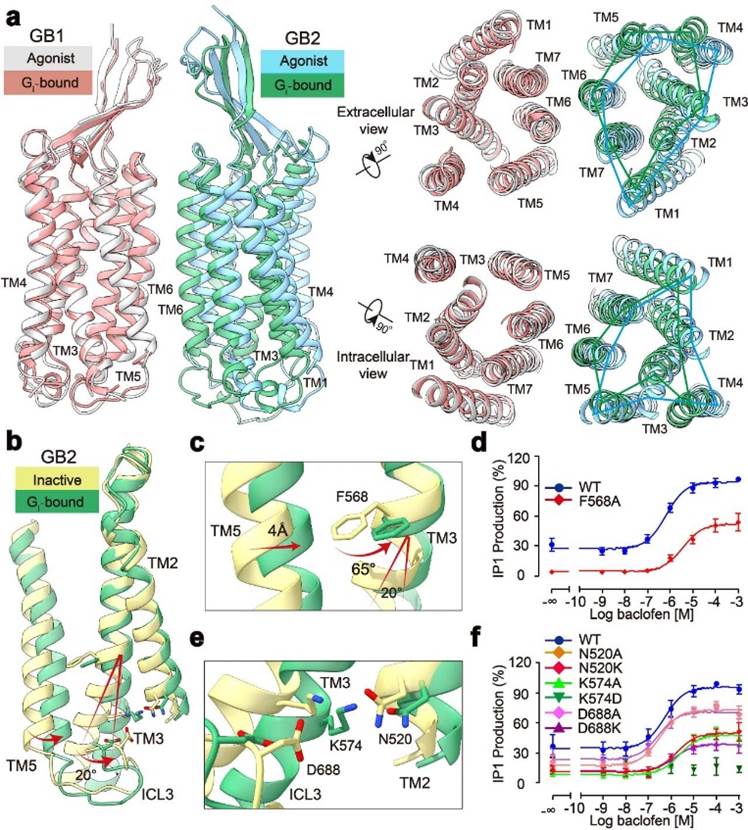
Figure 2: Activation mechanism of GABAB receptor with asymmetric changes
GABAB receptors are mainly coupled to Gi proteins through three intracellular loops (ICLs): the longer ICL2 mainly binds to α 5, α N, β 2- β 3 on G proteins, occupying about 80% of the interaction interface; The shorter ICL1 and ICL3 mainly interact with the α 5 terminal amino acids of G protein, stabilizing the binding of G protein (Figure 3a, b). In addition, studies have found that GABAB receptors selectively recognize Gi/o subtype G protein because the H5.23 site of Gi/o is the cysteine of the small side chain, while the tyrosine of the large side chain at the corresponding position of G proteins such as Gs and Gq has potential steric hindrance with GABAB receptors (Figure 3c). Functional experiment analysis found that when the cysteine at H5.23 site was mutated into tryptophan with a large side chain, its ability to bind to G protein significantly decreased, while when it was mutated into alanine with a small side chain, its ability to bind to G protein did not change (Figure 3d, e).
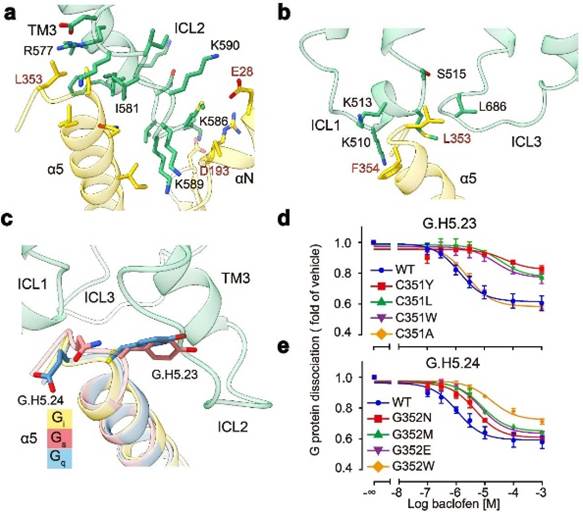
Figure 3: Molecular mechanism of GABAB receptor selective coupling with Gi protein
This work fully elucidates the asymmetric activation mechanism of C-class heterodimeric GABAB receptors: in the inactive state, the VFT regions of both subunits of GABAB receptors are in an open state, and the TMD region adopts the TM3-TM5/TM3-TM5 interaction interface; After the agonist binds to the positive pocket of GB1-VFT, it induces the closure of GB1-VFT, leading to the rotation of GB2-VFT and further conduction from the neck region to the rearrangement of TMD, forming a new TM6/TM6 interface; The binding of PAM and G protein further induces the rotation of TMD in GB2, forming a more stable TM6/TM6 interaction interface; In addition, the fine conformational changes of residues such as GB2 subunit F568 cause the intracellular end of TM3/TM5 to shift outward, forming shallow pockets for selective coupling of Gi/o proteins within the three intracellular loops, ultimately activating downstream signaling pathways (Figure 4).
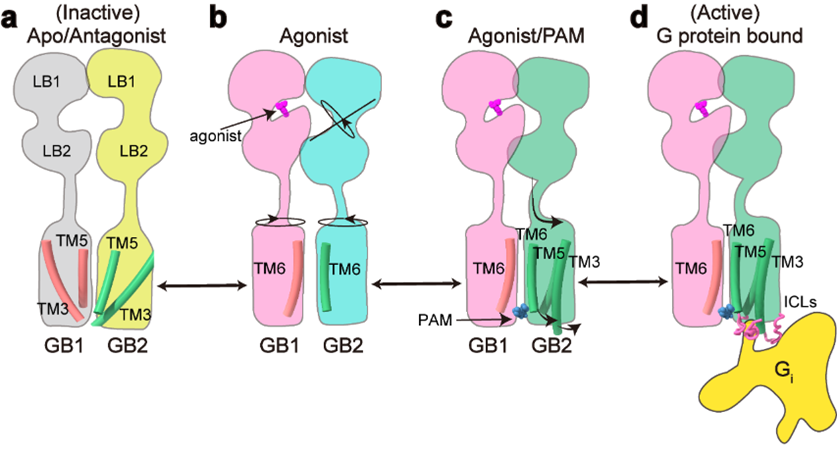
Figure 4: Signal transduction model of GABAB receptor activation
This study is another major breakthrough in the research of C-class GPCR signal transduction mechanism by the Zhejiang University Huazhong University of Science and Technology Joint Laboratory of Cell Signal Transduction. This work further validates the activation patterns of various functional domain conformational modulation of different subunits in C-class GPCR dimers previously discovered by Jianfeng Liu's research group, such as metabotropic glutamate receptors (Nat Chem Biol., 2015; PNAS., 2011; J Biol Chem., 2006), GABAB receptors (Cell Res., 2020; NatCommun., 2019; EMBO J., 2008), and calcium sensitive receptors (PNAS., 2020). The first authors of this study are Cangsong Shen, a doctoral student jointly trained by Huazhong University of Science and Technology and the School of Medicine of Zhejiang University, Chunyou Mao, a researcher at the Run Run Shaw Hospital affiliated with the School of Medicine of Zhejiang University, Chanjuan Xu, a teacher at Huazhong University of Science and Technology, and Nan Jin, a doctoral student. Professor Jianfeng Liu from the School of Life Sciences and Technology at Huazhong University of Science and Technology, Researcher Yan Zhang from the School of Medicine at Zhejiang University, and Professor PIN JP from the French Institute of Functional Genetics are co corresponding authors. The Protein Platform and Cryo Electron Microscopy Center of Zhejiang University School of Medicine provided support for sample preparation and data collection in this study. This work was also assisted by Professor Zhong Chen from Zhejiang University of Traditional Chinese Medicine, Professor Tingjun Hou from Zhejiang University, and Professor Rondard P from the French Institute of Functional Genetics.
